Training School 1 | Integrating experimental and computational approaches in protein self-assembly
Venue: Københavns Biocenter, Biologisk Institut, Copenhagen, Denmark
Dates: Nov 13-16, 2023
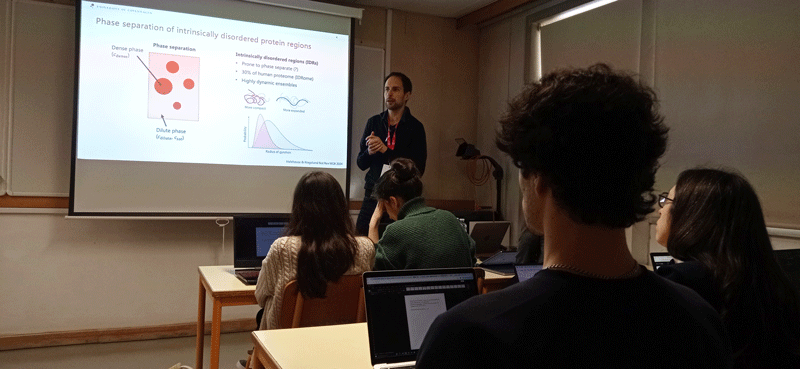
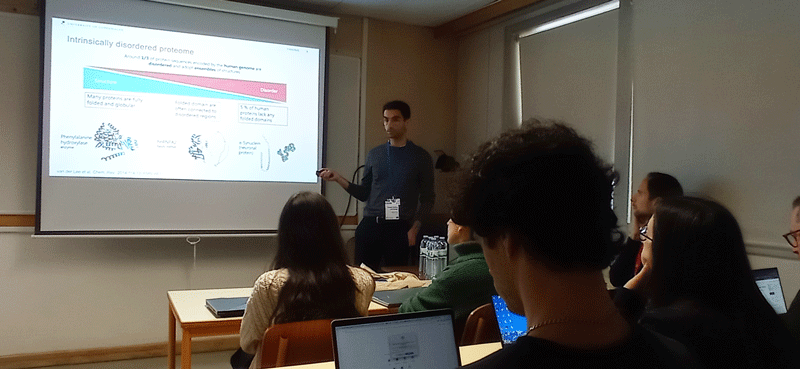
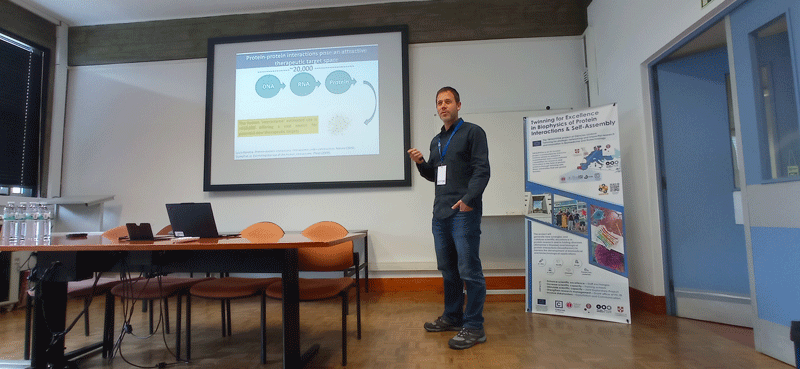
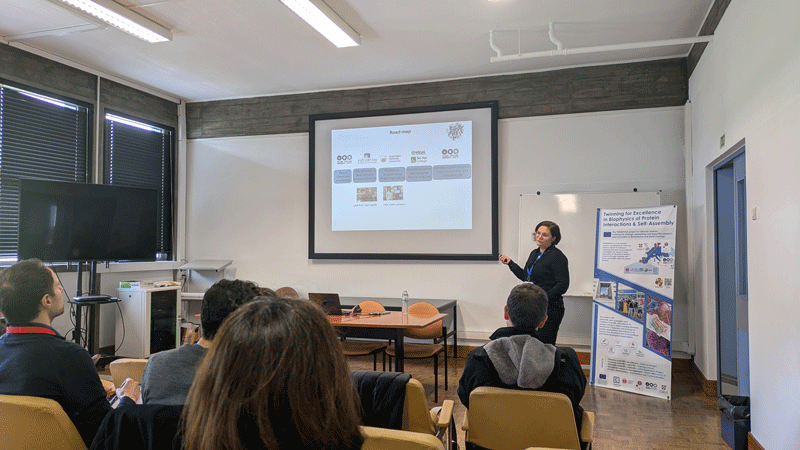
Objective: Introduce the network to the advanced experimental and computational techniques applied the study of proteins, their interactions, and the relationships to function and disease. Participants will acquire knowledge and practical utility of NMR and SAXS to study dynamical proteins and their assemblies; key steps involved in running molecular dynamics simulations; integration of experiments and simulations, including knowledge of key approaches and software; knowledge and data analysis of computational methods for predicting variant effects (disease mutations) using protein structure and sequence; understand the role of protein quality control in disease and experimental methods to study these; knowledge and basic overview of how multiplexed assays of variant effects can be used to study protein structure-function relationships at scale.